Research Overview
The cancer genome informatics group at Rutgers Cancer Institute seeks to identify the origins and implications of genetic and epigenetic abnormalities in different cancer types using genomics and computational approaches. We develop new methods and work in close collaboration with bench scientists and clinicians. To learn more about our lab, and employment and collaborative opportunities, please visit www.sjdlab.org.
Genomic Instability in Cancer
We have recently shown that nuclear localization, DNA replication timing and genomic context play critical roles in shaping the landscape of amplifications, deletions, point mutations, and loss of heterozygosity in cancer genomes. Our findings provide insights into different mutagenic processes, and has implications for identifying driver and passenger mutations in the cancer genomes.
Pedersen et al. Nucleic Acids Res. 2013
Liu et al. Nature Comm. 2013
Podlaha et al. Trends Genet. 2012
De et al. Nature Biotech.. 2011
De et al. Nature Str. Mol. Biol. 2011
Regulatory Alterations in Cancer
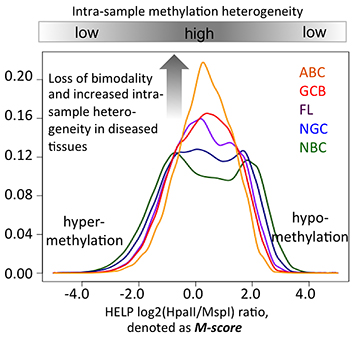
Genomic and epigenetic abnormalities leading to deregulated oncogenic pathways are found in many types of cancer. We found a novel signature of accelerated somatic evolution in many types of cancer, which is marked by significant excess of somatic mutations in a genomic region in multiple cancer genomes. The signature is frequently associated with non-coding regulatory changes leading to deregulation of oncogenic pathways and adverse clinical outcome. In another project, we found that epigenetic heterogeneity in B cell lymphoma is increases markedly with disease aggressiveness, and is associated with unfavorable clinical outcome. Our findings provide mechanistic insights into the biology of regulatory alterations in cancer.
Smith KS et al. Nucleic Acids Res. 2015
Shaknovich, De, Michor. Biochim Biophys Acta. 2014
De, Shaknovich, Riester et al. PLoS Genetics. 2013
Shaknovich et al. Blood. 2011
Evolutionary Dynamics of Cancer

We use a combination of computational, genomics, and mathematical tools to study evolutionary dynamics of cancer, including order of mutation events and emergence of resistance. We found that not only the driver mutations are important, but the order in which they arise is also important. Although there is no obligatory order of events, we found that loss of PTEN is the most common first event and is associated with basal-like subtype, whereas in the majority of luminal tumors, mutation of TP53 occurs first and mutant PIK3CA is rarely detected. Furthermore, resistant mutations are often present at low frequency during treatment. Our results have important implications for the design of chemopreventive and therapeutic interventions in this high-risk patient population.
Martins et al. Cancer Discovery. 2012 (cover story)
Smith et al. Bioinformatics. 2016
Hintzsche J et al. JAMIA 2016
Somatic Mutations in Benign Human Tissues

We use a combination of computational, genomics, and mathematical tools to study evolutionary dynamics of cancer, including order of mutation events and emergence of resistance. We found that not only the driver mutations are important, but the order in which they arise is also important. Although there is no obligatory order of events, we found that loss of PTEN is the most common first event and is associated with basal-like subtype, whereas in the majority of luminal tumors, mutation of TP53 occurs first and mutant PIK3CA is rarely detected. Furthermore, resistant mutations are often present at low frequency during treatment. Our results have important implications for the design of chemopreventive and therapeutic interventions in this high-risk patient population.
Martins et al. Cancer Discovery. 2012 (cover story)
Smith et al. Bioinformatics. 2016

